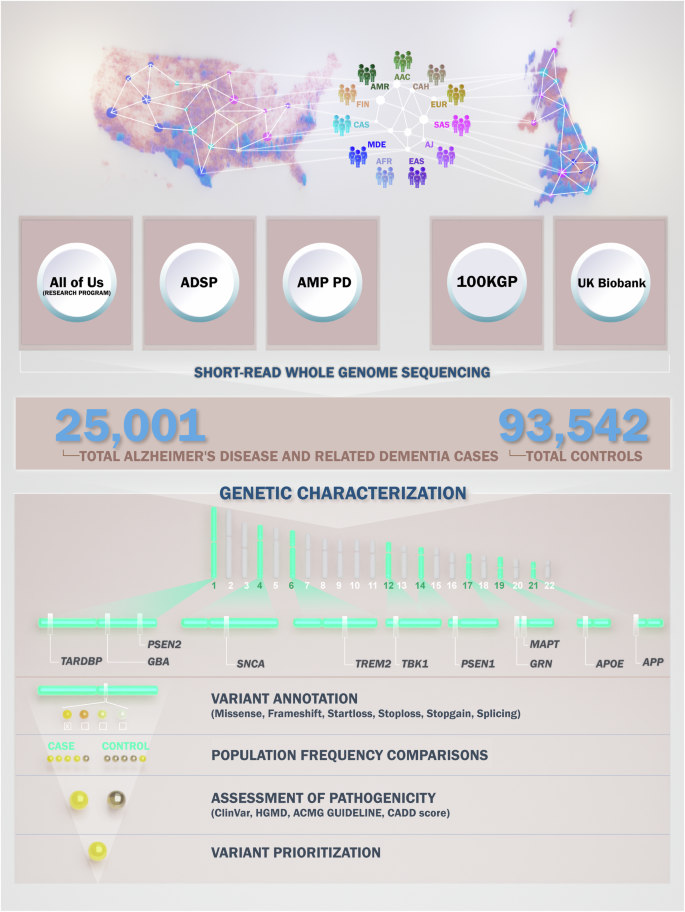
Abstract: Alzheimer’s disease and related dementias (AD/ADRDs) affect over 55 million people worldwide, with cases expected to nearly triple by 2050. While most genetic studies have focused on European populations, this new large-scale research takes a groundbreaking multi-ancestry approach, analyzing over 25,000 cases and 93,000 controls across 11 ancestries using data from major biobanks such as All of Us, UK Biobank, ADSP, and more. The study identified both known and novel variants in key dementia-related genes like APP, PSEN1, PSEN2, TREM2, MAPT, GRN, GBA1, and APOE. Crucially, it highlights how genetic architecture varies across ancestries, reshaping our understanding of global risk, resilience, and therapeutic targets for Alzheimer’s and other dementias.
The researchers also uncovered protective and disease-modifying variants that may delay or lessen the severity of dementia, particularly in APOE ε4 carriers, the group most genetically vulnerable to AD. Interestingly, several variants long thought to be “disease-causing” may have incomplete penetrance, appearing in healthy individuals as well. These findings stress the importance of studying diverse populations, since certain variants showed significance only in African, South Asian, or admixed ancestries. Beyond mapping genetic risk, the team launched an interactive platform to explore these results, empowering scientists worldwide to accelerate dementia research and guide precision medicine.
Explore the full research here: Biobank-scale genetic characterization of Alzheimer’s disease and related dementias across diverse ancestries | Nature Communications.