Microbiomes are dynamic and complex systems consisting of – bacteria, archaea, fungi, algae, protists, and viruses. Microbiome research started back in the 17th century and rapidly evolved in subsequent years. However, the biobanking infrastructure is currently not prepared for the biobanking of microbiomes. As it stands today, biobanks primarily focus on particular aspects of a microbiome and preserve them individually as snapshots instead of as a part of a bigger ecosystem.
Due to environmental and climatic variations, we are losing some ecologically and industrially relevant microbial community components. Microbiome preservation is crucial for future cultivation and to study the effects of ecological and climatic variations on community functionality and shift with time. Although methods for pure culture preservation are almost optimized, microbiome preservation techniques remain an unsolved challenge for microbiologists due to technical and physiological constraints.
This paper explores the different aspects of research in microbiome biobanking and answers tertiary questions around the field, such as the necessity of the area, as well as possible methodologies of how to implement them.
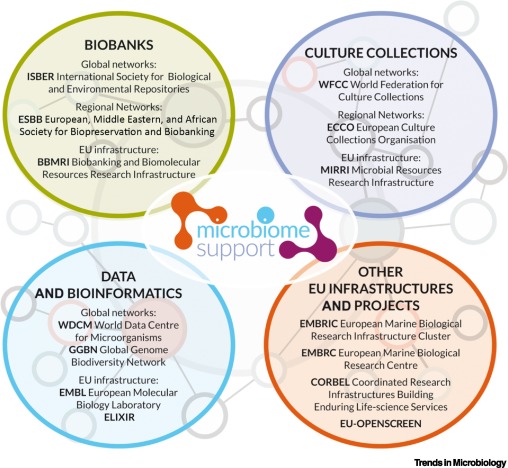
The Current European and International Landscape Underpinning Microbiome Research Is Fragmented.
Preservation and Storage:
The challenges of preserving microbiome samples are immense. Removing a single critical microbial component at the functional level due to applying a non-optimized storage approach could irreversibly affect the system’s integrity.
Bell quoted Adams ‘If you try and take a cat apart to see how it works, the first thing you have on your hands is a non-working cat,’ which is a crucial issue to conserve a microbiome sample.
When considering microbiome preservation, two essential questions need to be answered:
- What should be preserved, and
- What is the best way of preserving it?
Capacity is one significant challenge to this approach. The question of how much sample is required to be representative of the microbiome in question becomes critical.
Cryopreservation has been the ‘gold standard’ for microbial storage since the 1960s. But in the microbiome system of multiple components, if cryopreservation is not applied optimally, it will result in unintended selective pressures on the community.
Assessing Success and Quality:
A variety of metagenomics to transcriptomics has been used to assess the microbiome concerning both its construction and functionality. These approaches could evaluate the success of preservation and the storage regime, but each has its limitations. Tests should be undertaken before preservation/storage to characterize the microbiome, and one post preservation to ensure the compositional and functional integrity.
Summary Recommendations and the Way Forward:
The question of why and what should be conserved has to be addressed in detail considering scientific, economic, social, and environmental perspectives. A prioritized list is required to focus the efforts and achieve advancements, considering the diversity and complexity of microbiomes across environments.
The most significant technological bottleneck is the development of optimized methodologies for preserving microbiomes and assessing preservations’ success in maintaining the composition and functionality of microbiomes.
The evident complementarity between culture collections and biobanks necessitates an approach to enable both works together to ensure that this critical microbiome research field has adequate support. This will require identifying infrastructural overlaps to gauge what is needed and what is available/missing within the EU and beyond.
How can OpenSpecimen Help?
Managing the data related to microbes, the primary component of the microbiomes, is equally essential. OpenSpecimen helps in managing the data of the complete life cycle of microbes, including preservation and storage. The diagnostic division of Clinical Bacteriology and Mycology of Prof. Adrian Egli in University Hospital Basel, Switzerland, uses OpenSpecimen regularly to manage their longitudinal bacterial strain collections from routine diagnostics. This includes samples for quality measurements or outbreak management. Besides, a series of clinical research trials are also managed by OpenSpecimen, e.g.,
- A study exploring the skin microbiome in patients with Graft Versus Host Diseases (GVHD) including swab and skin-tissue samples collection to study skin conditions
- A study to explore community-acquired pneumonia and associated sepsis, which involves the collection of different types of specimens to study changes during severe community-acquired pneumonia and subsequent development of sepsis.
For more details, refer to the case study of the University Hospital Basel, Switzerland.
Acknowledgments
This paper is an output from the EU project MicrobiomeSupport. For more details, refer to the original paper here.
References:
- https://en.wikipedia.org/wiki/Microbiome
- https://www.youtube.com/watch?v=VzPD009qTN4&feature=youtu.be
- https://en.wikipedia.org/wiki/Microbiological_culture
- https://www.sciencedirect.com/science/article/pii/S0966842X20301888#bb0015
Written by: Divya Prabhu, member of domain staff at OpenSpecimen.
For more details email [email protected]