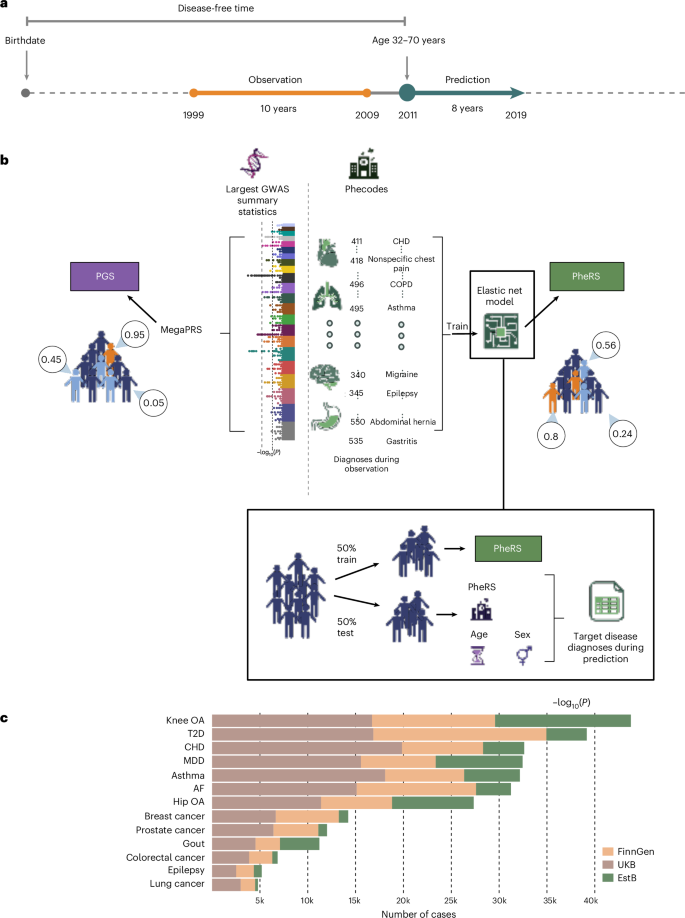
Abstract: A new study published in Nature Genetics (2025) explored how electronic health record (EHR)-based risk scores (PheRS) can complement polygenic scores (PGS) to improve disease prediction. Researchers analyzed data from over 845,000 individuals across three major biobanks, FinnGen, UK Biobank, and Estonian Biobank, tracking the onset of 13 common diseases, including type 2 diabetes, coronary heart disease, asthma, depression, and several cancers. The study found that PheRS, built from patients’ past diagnoses, were strongly associated with disease risk and generalized well across healthcare systems, even without retraining. Interestingly, PheRS captured largely independent information from PGS, making them a valuable addition rather than a replacement.
The results highlight that combining PheRS with PGS significantly improved disease onset prediction for 8 out of 13 diseases, showing particular strength for conditions like type 2 diabetes, gout, asthma, and depression. PheRS also proved more effective than PGS in identifying individuals at the very highest risk in several diseases. Unlike genetic data, EHRs are routinely collected, making them cost-effective and widely accessible. While genetic risk scores are powerful, their limited generalizability across ancestries poses challenges. This study shows that integrating clinical histories with genetic information provides a more robust, equitable, and scalable way to predict disease, potentially shaping the future of personalized medicine.
Read the full research here: Cross-biobank generalizability and accuracy of electronic health record-based predictors compared to polygenic scores | Nature Genetics